4 Example Krill MSE
Showcasing the application of a Management Strategy Evaluation to the Krill fishery using the openMSE tool
4.1 Introduction
4.2 Set-up MSE
4.2.1 Build the base-case Operating Model
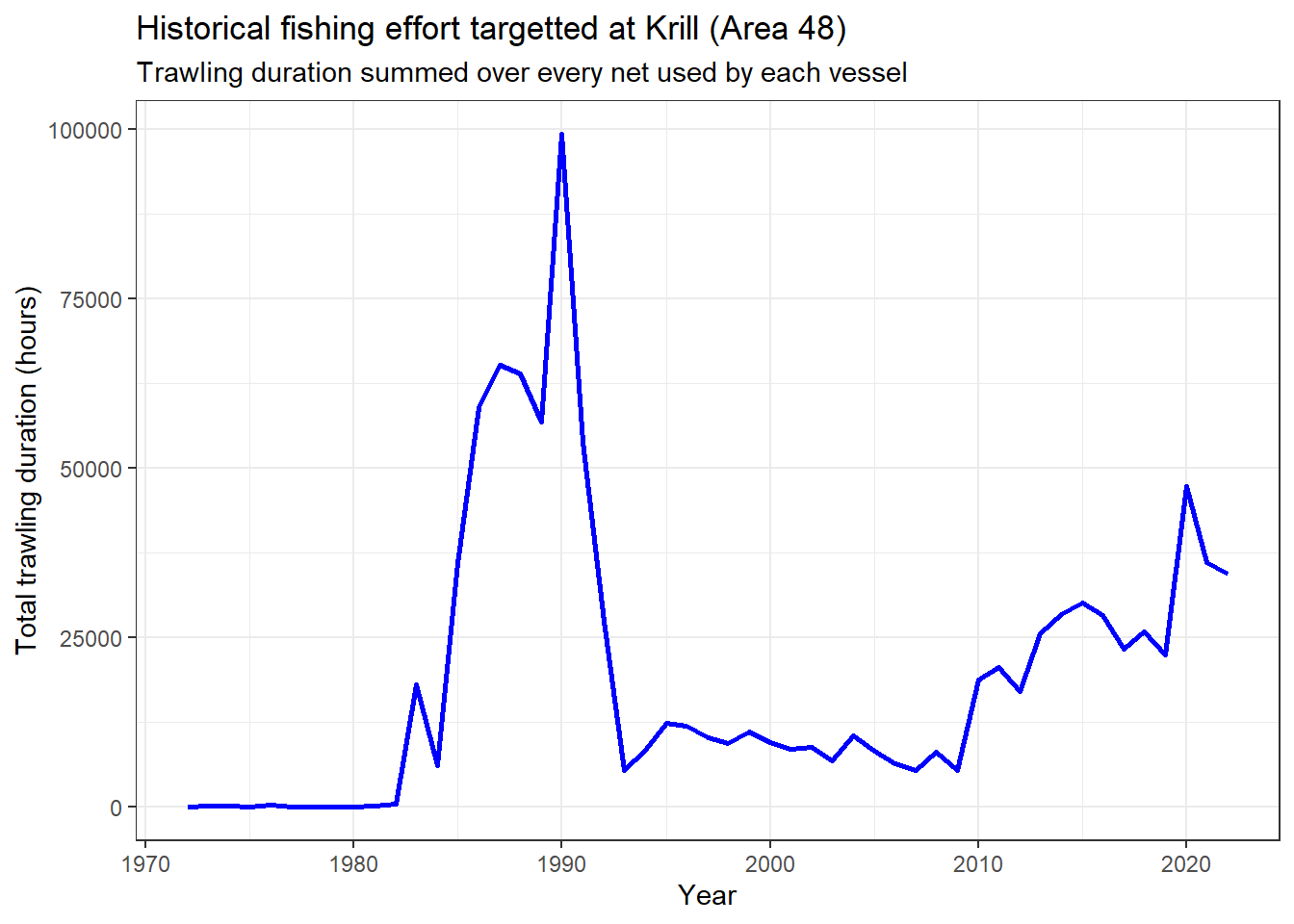
Compute time-series of yearly total fishing effort targeted at Krill in terms of trawling duration in hours (summed over each net in each vessel) in the whole Statistical Area 48 (source: CCAMLR Secretariat 2023).
# filter effort for krill in all subareas of 48
effort_krill_48 <- effort_ccamlr |>
filter(
target_species_code == "KRI",
asd_code %in% as.character(c(48, 481:486))
)
yearly_trawl_hrs_krill_48 <- effort_krill_48 |>
group_by(year) |>
summarise(trawl_hrs = sum(trawl_duration_hours, na.rm = TRUE))
ggplot(yearly_trawl_hrs_krill_48) +
geom_line(aes(x = year, y = trawl_hrs), col = "blue", linewidth = 1) +
labs(
x = "Year",
y = "Total trawling duration (hours)",
title = "Historical fishing effort targetted at Krill (Area 48)",
subtitle = "Trawling duration summed over every net used by each vessel"
)
The base-case OM is specified next, taking the PR estimates and maturity ogive defined under scenario scn-2 of the openMSE-Grym approximation analysis presented in Part 1 of this project (Table 2.3). This scenario comprises random draws of natural mortality rate \(M\) that are closest to the range of estimates presented in Pakhomov (1995), and maturity ogive estimates recommended in Maschette et al. (2021).
[Add main differences to OMs specified in openMSE-Gryn approximation]
# Selecting scenario "scn-2"
krill_pars <- grym_setups |>
filter(scenario_id == "scn-2")
krill_base_OM <- build_krill_OM(
om_name = "krill_base_case",
maxage = last(unlist(krill_pars$Ages)),
nyears = nrow(yearly_trawl_hrs_krill_48),
LenCV = c(0.05, 0.08), # low variation in length-at-age
effort = yearly_trawl_hrs_krill_48$trawl_hrs,
qinc = c(0.0), # no increase in fishing efficiency
M_draws = krill_pars$prRecruitPars$`PR-emm21`$M,
RCV_draws = krill_pars$prRecruitPars$`PR-emm21`$CV,
n_iter = krill_pars$n_iter,
proj_yrs = krill_pars$n.years,
maxF = krill_pars$Fmax,
seed = 101
)
write_rds(krill_base_OM, "inputs/krill_base_OM.rds")Next we render the OM report documenting all the parameter values specified under the base-case OM, along with the rationale behind their selection. The base-case OM report is available in Appendix B.
# Modified versions of the MSEtool scripts solve bugs, in addition to adding the
# inclusion and better integration in another document as an embedded html
source("../MSEtool_fixes/OM_init_doc_dmp.R")
source("../MSEtool_fixes/OM_plots_dmp.R")
OMdoc(
OM = krill_base_OM,
openFile = FALSE,
rmd.source = "OM_krill_mse_base.rmd",
bib_file = "../references.json",
html_theme = "lumen",
inc_title = FALSE
)4.2.2 Specify Management Procedures
[FO->] The choice of MPs applicable to the krill stock is conditioned by their data requirements. The krill fishery can be classified as data-limited, with mostly time-series of catches and effort available on a consistent basis. There are frequent scientific surveys but only cover specific sub-areas of the management region [<-FO]
The following assumptions are required in terms of data availability in order to apply the MPs described in Table 4.1:
Time-series of catch at length can be estimated using length-frequency data collected from observer sampling.
Time-series of fishery-dependent CPUE are obtainable from annual catch and effort data. These CPUE estimates can be standardized to provide a suitable relative index of abundance.
Prescribed MSY-based reference points can be either calculated (e.g. the method by Martell and Froese, 2012) or specified via expert elicitation.
Code
MPs <- read_xlsx(
"krill applicable MPs.xlsx",
sheet = "picked_MPs_description"
)
MPs_names <- MPs$Name
MPs |>
flextable() |>
add_header_row(
values = c("General Category", "Name", "Description", "Data Requirements"),
colwidths = c(1, 1, 1, 5)
) |>
merge_v( j = 1:3, part = "header") |>
merge_v( j = 1, part = "body") |>
align(i = 1:2, align = "center", part = "header") |>
align(j = 4:8, align = "center", part = "body") |>
footnote(
#i = ~select(Name, starts_with("Fdem")), j = 8,
i = ~str_detect(Name, "Fdem"),
j = "Prescribed F_MSY/M",
value = as_paragraph(
"Requirement according to documentation and input validation in source code. However FMSY_M is not used anywhere in `Fdem_` functions... so assuming requirement is non-valid."
),
ref_symbols = "ǂ") |>
bg(j = ~`Catch series`, i = ~str_detect(`Catch series`, "X"), bg = "#E1F5FE", part = "body") |>
bg(j = ~`Index series`, i = ~str_detect(`Index series`, "X"), bg = "#E1F5FE", part = "body") |>
bg(j = ~`Catch@length series`, i = ~str_detect(`Catch@length series`, "X"), bg = "#E1F5FE", part = "body") |>
bg(j = ~`Life history parameters`, i = ~str_detect(`Life history parameters`, "X"), bg = "#E1F5FE", part = "body") |>
bg(j = ~`Prescribed F_MSY/M`, i = ~str_detect(`Prescribed F_MSY/M`, "X"), bg = "#E1F5FE", part = "body") |>
bg(bg = "#FFFFFF", part = "header") |>
set_table_properties(
layout = "autofit",
opts_word = list(split = TRUE),
opts_html = list(scroll = list(height = "800px"))
) General Category | Name | Description | Data Requirements | ||||
|---|---|---|---|---|---|---|---|
Catch series | Index series | Catch@length series | Life history parameters | Prescribed F_MSY/M | |||
Status-quo | AvC | Constant TAC over projection period based on average historical catch. Represents the "status quo" management option. | X | ||||
Stabilisation | Islope1 | TAC ajusted incrementally to maintain stable CPUE or abundance index. Based on average recent catch (last 5 years) and the trajectory of index of abundance over the past 5 years. | X | X | |||
Islope4 | Similar to Islope1, but more precautionary. Uses a fraction (0.6) of average recent catch and forces slower changes in TAC in response to changes in cpue/index trajectory | X | X | ||||
LstepCC1 | TAC adjusted stepwise in relation to recent changes in mean length of recent catches. Takes the average historical catch (last 5 years) and applies a (upwards or downwards) step change depending on the ratio between mean length in recent catch (passt 5 years) and mean length in historical catches (last 10 years). | X | X | ||||
LstepCC4 | Similar toLstepCC1 but more precautionary. Uses a fraction (0.7) of average historical catch | X | X | ||||
Target-based | Itarget1 | TAC adjusted to achieve a target CPUE/index. Based on average historical catch (last 5 years) and adjustment term expressing deviations between historical, recent and target indices. Target index defined as 1.5 of average index over the last 10 years of historical period. Recent average index calculated from past 5 years. | X | X | |||
Itarget4 | Similar to Itarget1 but more precautionary. Uses a fraction (0.7) of average historical catch and target index is higher (2.5 of average historical index) | X | X | ||||
Ltarget1 | TAC incrementally adjusted to reach a target mean length in catches. Based on average historical catch (last 5 years) and adjustment term expressing deviations between historical, recent and target mean lengths. Target mean length defined as 1.05 of average length over the last 10 years of historical period. Recent average length in catch calculated from past 5 years | X | X | ||||
Ltarget4 | Similar to Ltarget1 but more precautionary. Uses a fraction (0.8) of average historical catch and target mean length is higher (1.15 of historical mean length) | X | X | ||||
L95target | Same as Ltarget1 but here the target mean length is based on the length at maturity rather than an arbitrary multiplicative | X | X | X | |||
L95target4 | As L95target but more precautionary. Uses a fraction (0.8) of average historical catch | X | X | X | |||
MSY-based | Fdem_ML | Annual TAC set for harvesting at MSY levels based on F_MSY. F_MSY calculated as r/2, where r Is the intrinsic rate of population growth. r is estimated from life history parameters. Current abundance estimated from mean length | X | X | X | Xǂ | |
Fdem_ML_25 | As Fdem_ML but with a precautionary buffer of 0.25 of the recommended TAC | X | X | X | Xǂ | ||
Fdem_ML_50 | As Fdem_ML but with a precautionary buffer of 0.50 of the recommended TAC | X | X | X | Xǂ | ||
Fdem_ML_75 | As Fdem_ML but with a precautionary buffer of 0.75 of the recommended TAC | X | X | X | Xǂ | ||
SPMSY | Sets TAC to achieve harvesting at MSY. MSY estimated from Martell and Froese surplus production model. Stock trajectories estimated based on trends in catch data and life-history information | X | X | ||||
SPMSY_25 | As SPMSY but with a precautionary buffer of 0.25 of the recommended TAC | X | X | ||||
SPMSY_50 | As SPMSY but with a precautionary buffer of 0.50 of the recommended TAC | X | X | ||||
SPMSY_75 | As SPMSY but with a precautionary buffer of 0.75 of the recommended TAC | X | X | ||||
DD | Sets TAC to achieve exploitation at UMSY, i.e. the harvest rate (the fraction of population being harvested every year) that will produce MSY. UMSY estimated from a simple delay-difference model (described in chap 9 of Hilborn and Walters, 1992) using time-series of catch and index of abundance | X | X | X | |||
DD_25 | As DD but with a precautionary buffer of 0.25 of the recommended TAC | X | X | X | |||
DD_50 | As DD but with a precautionary buffer of 0.50 of the recommended TAC | X | X | X | |||
DD_75 | As DD but with a precautionary buffer of 0.75 of the recommended TAC | X | X | X | |||
DD4010 | Similar to DD but superimposing a 40-10 HCR to constrain TAC recommendation as a function of depletion (B/B0). 40-10 rule specifies that the stock is not fished if depletion < 10% (i.e. TAC = 0), while TAC is set at UMSY levels (as DD) if depletion > 40%. If depletion is between 10 and 40%, TAC follows a linear increase from 0 to 100% UMSY levels. | X | X | X | |||
DD4010_25 | As DD4010 but with a precautionary buffer of 0.25 of the recommended TAC | X | X | X | |||
DD4010_50 | As DD4010 but with a precautionary buffer of 0.50 of the recommended TAC | X | X | X | |||
DD4010_75 | As DD4010 but with a precautionary buffer of 0.75 of the recommended TAC | X | X | X | |||
Yield-per-recruit | BK_ML | Annual TAC based on F_Max. F_max derived using length at first capture relative to L_inf and vB growth parameter K. Current vulnerable abundance estimated from recent catches and F, with F estimated from mean length | X | X | X | ||
BK_ML_25 | As BK_ML but with a precautionary buffer of 0.25 of the recommended TAC | X | X | X | |||
BK_ML_50 | As BK_ML but with a precautionary buffer of 0.50 of the recommended TAC | X | X | X | |||
BK_ML_75 | As BK_ML but with a precautionary buffer of 0.75 of the recommended TAC | X | X | X | |||
YPR_ML | TAC set to achieve harvest at F_0.1. Yield-per-recruit curve calculated from an age-structured model at equilibrium with life-history and selectivity parameters sampled from OM. Current abundance inferred from an estimate of Z calculated from mean length in catches and life-history parameters | X | X | X | |||
ǂRequirement according to documentation and input validation in source code. However FMSY_M is not used anywhere in `Fdem_` functions... so assuming requirement is non-valid. | |||||||
4.2.3 Set Performance Metrics
[Add details on newly introduced performance metrics]
Code
esc_metric <- ESC(short_krill_mse)
pd_metric <- PD(short_krill_mse)
ssb_fvu_metric <- SSB_FvU(short_krill_mse, Ref = 0.70)
yrc_metric <- YRC(short_krill_mse)
lty_metric <- LTY(short_krill_mse)
aavy_metric <- AAVY(short_krill_mse)
PMs <- tribble(
~ Category, ~ Name, ~ Description, ~ Detail,
"Spawning Biomass", "ESC", esc_metric@Name, esc_metric@Caption,
"Spawning Biomass", "PD", pd_metric@Name, pd_metric@Caption,
"Spawning Biomass", "SSB_FvU", ssb_fvu_metric@Name, ssb_fvu_metric@Caption,
"Yield", "YRC", yrc_metric@Name, yrc_metric@Caption,
"Yield", "LTY", lty_metric@Name, lty_metric@Caption,
"Yield", "AAVY", aavy_metric@Name, aavy_metric@Caption
)
PMs |>
flextable() |>
merge_v(j = ~Category) |>
fix_border_issues() |>
footnote(
i = ~ str_detect(Name, "ESC|PD"),
j = "Name",
value = as_paragraph("CCAMLR's default metrics"), ref_symbols = "ǂ")Category | Name | Description | Detail |
|---|---|---|---|
Spawning Biomass | ESCǂ | Escapement: Final Spawning Biomass relative to Virgin Spawning Biomass | median(SSB at Year 20)/median(SSB0) |
PDǂ | Depletion: Projected Spawning Biomass relative to Virgin Spawning Biomass | Prob. min(SSB) < 0.2 SSB0 (Years 1 - 20) | |
SSB_FvU | Proportion of projection years with precautionary levels of exploitation | Proportion of years with SSB > 0.7 Unfished SSB (Years 1-20) | |
Yield | YRC | Projected Average Yield relative to Historical Average Yield | Prob. Average Projected Yield (Years 11-20) > Average Historical Yield (-9-0) |
LTY | Average Yield relative to Reference Yield (Years 11-20) | Prob. Yield > 0.5 Ref. Yield (Years 11-20) | |
AAVY | Average Annual Variability in Yield (Years 1-20) | Prob. AAVY < 20% (Years 1-20) | |
ǂCCAMLR's default metrics | |||
4.2.4 Generate Operating models for robustness testing
Considering five cases for testing the robustness of results under the base-case MSE (Table 4.3):
Sensitivity to higher variability in length-at-age (parameter LenCV)
Increase in catchability for the projection period (parameter qinc), i.e. fishing becomes more efficient and one unit of effort will lead to higher fishing mortality. Setting it to 1% annual increase (resulting in a 20% increase in fishing efficiency after a 20 year projection), to express advances in e.g. gear technology, generalized use of continuous fishing systems, etc.
Presence of persistent bias in reported catches (parameter Cbiascv), e.g. due to inconsistencies in the estimation of green weight. A CV of 5% implies that 95% of simulations are between 90% and 110% of the true simulated catches.
Presence of hyperstability in the estimation of abundance (parameter beta), i.e. acknowledging that estimated CPUE indices may decrease slower than the true actual abundance. This phenomenon may occur, for instance, if fishing vessels tend to move uni-directionally across sub-areas during each fishing season while krill spatial distribution is uniform.
Presence of persistent bias in estimates of natural mortality rate (parameter Mbiascv), e.g. as a by-product of the Proportional Recruitment method. A CV of 5% implies that 95% of simulations are between 90% and 110% of the true simulated natural mortality.
Code
rbstn_vals <- list(
LenCV = c(0.15, 0.20),
qinc = c(0.01, 0.01),
Cbiascv = 0.05,
beta = c(0.8, 0.9),
Mbiascv = 0.05
)
rbstn_tbl <- tribble(
~"Robustness Test ID", ~"OM Parameter", ~"Base-case", ~"Robustness",
"Length@Age", "LenCV", "[0.05, 0.08]", glue("[{toString(rbstn_vals$LenCV)}]"),
"Fishing-efficiency", "qinc", "[0, 0]", glue("[{toString(rbstn_vals$qinc)}]"),
"Catch-bias", "Cbiascv", "0", glue("{toString(rbstn_vals$Cbiascv)}"),
"Hyperstability", "beta", "[0, 0]", glue("[{toString(rbstn_vals$beta)}]"),
"M-bias", "Mbiascv", "0", glue("{toString(rbstn_vals$Mbiascv)}"),
)
flextable(rbstn_tbl) |>
set_table_properties(width = 0.75, layout = "autofit")Robustness Test ID | OM Parameter | Base-case | Robustness |
|---|---|---|---|
Length@Age | LenCV | [0.05, 0.08] | [0.15, 0.2] |
Fishing-efficiency | qinc | [0, 0] | [0.01, 0.01] |
Catch-bias | Cbiascv | 0 | 0.05 |
Hyperstability | beta | [0, 0] | [0.8, 0.9] |
M-bias | Mbiascv | 0 | 0.05 |
Code
temp_base_OM <- krill_base_OM
robustness_OMs <- imap(rbstn_vals, \(x, y){
slot(temp_base_OM, name = y) <- x
temp_base_OM
})
names(robustness_OMs) <- glue("rbst_{names(robustness_OMs)}_OM")
write_rds(robustness_OMs, "inputs/robustness_OMs.rds")4.3 Run MSE Simulations
4.3.1 Base-case scenario
#' -------------------------------------------
#' Run Simulations for Base-Case MSE
#' ------------------------------------------
# WARNING: Simulations are actually run on a separate file
# 'mse_application_simulations.r'. Code provided below is just for reference
# Running this code in the quarto document is not advisable
# following recommended in documentation that Length bins should be specified when running in 'sac1 parallel mode.
# Bins based on length frequency samples published in te Krill Fishery report (2023)
krill_base_OM@cpars$CAL_bins <- seq(0, krill_base_OM@Linf[2] + 5, by = 2)
tic()
krill_mse <- runMSE(OM = krill_base_OM, MPs = MPs_names, parallel = "sac")
toc()
write_rds(
krill_mse,
file = fs::path(outputs_path, "krill_mse_basecase.rds"),
compress = "gz"
)4.3.2 Robustness scenario
# WARNING: Simulations are actually run on a separate file
# 'mse_application_simulations.r'. Code provided below is just for reference.
# Running this code in the quarto document is not advisable
#' -------------------------------------------
#' Run Simulations for robustness MSE
#' ------------------------------------------
robustness_mse <- robustness_OMs[4:5] |>
imap(\(x, y){
#x <- robustness_OMs$rbst_rbst_LenCV_OM_OM
#y <- "rbst_rbst_LenCV_OM_OM"
#x@nsim <- 100
cli::cli_h1("Starting MSE run for {y} @ {Sys.time()}")
x@cpars$CAL_bins <- seq(0, x@Linf[2] + 5, by = 2)
# run MSE for current scenario
tictoc::tic()
mse_output <- runMSE(
OM = x,
MPs = MPs_names,
parallel = "sac"
)
runtime <- tictoc::toc(quiet = TRUE)
# write out mse outputs
write_rds(
mse_output,
file = fs::path(outputs_path, glue::glue("krill_mse_{y}.rds")),
compress = "gz"
)
cli::cli_alert_success("Finished MSE for {y}: {runtime$callback_msg}")
return(mse_output)
})4.4 Results
Code
krill_base_mse <- read_rds("outputs/krill_mse_basecase.rds")
#krill_base_mse <- read_rds("outputs/krill_mse_basecase_mini.rds")
#krill_mse_prelim <- read_rds("outputs/krill_mse_base_outputs_preliminary.rds")
# Run PMs on MSE simulations
krill_base_PMs_outputs <- list(
PD = PD(krill_base_mse, Ref = 0.2),
ESC = ESC(krill_base_mse),
SSB_FvU = SSB_FvU(krill_base_mse, Ref = 0.70),
YRC = YRC(krill_base_mse),
LTY = LTY(krill_base_mse),
AAVY = AAVY(krill_base_mse)
)
write_rds(krill_base_PMs_outputs, "outputs/PM_results/krill_base_PMs_outputs.rds", "gz")
# Virgin Biomass SSB0
krill_base_ssb0 <- krill_base_mse@OM$SSB0
write_rds(krill_base_ssb0, "outputs/PM_results/krill_base_SSB0.rds", "gz")
# draws of SSB in final projection year, per MP
krill_base_ssbY <- krill_base_mse@SSB[, , krill_base_mse@proyears]
write_rds(krill_base_ssbY, "outputs/PM_results/krill_base_SSBY.rds", "gz")Code
# read in PM outputs
kb_pms <- read_rds("outputs/PM_results/krill_base_PMs_outputs.rds")
# extract PM summary
kb_pms_summ <- kb_pms |>
imap(\(x, y){
tibble(
MP = x@MPs,
{{y}} := x@Mean)
}) |>
reduce(full_join, by = "MP") |>
mutate(`1 - PD` = 1 - PD, .after = "PD")
# lookup table for MP names and position in PM outputs
MP_idx2name <- tibble::tibble(
mp = kb_pms$PD@MPs,
mp_idx = 1:length(kb_pms$PD@MPs)
)
# set colors for facet strips
MP_strips_theme <- ggh4x::strip_themed(
background_x = ggh4x::elem_list_rect(
fill = ifelse(kb_pms$PD@MPs == "AvC", col_status_quo, col_facet_strip_bg)
),
by_layer_x = FALSE
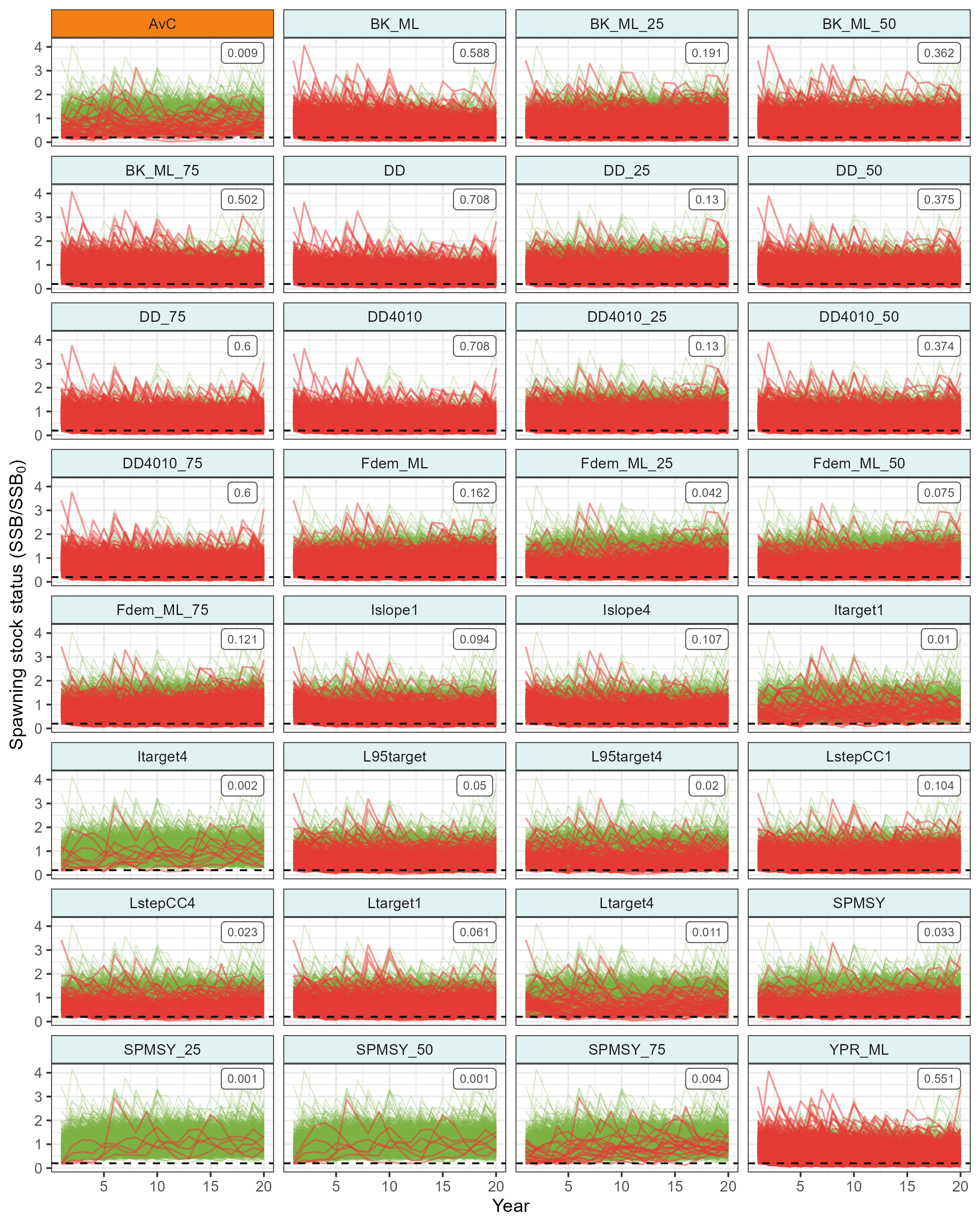
)Spawning Stock Status Projections
Code
krill_base_SSS <- reshape2::melt(
kb_pms$PD@Stat,
varnames = c("sim", "mp_idx", "year"),
value.name = "SSS") |>
dplyr::left_join(MP_idx2name, by = "mp_idx") |>
dplyr::mutate(
below_ref = if_else(min(SSS) < kb_pms$PD@Ref, TRUE, FALSE),
.by = c(sim, mp)
)
krill_base_PD_mtc <- tibble(
mp = kb_pms$PD@MPs,
PD = kb_pms$PD@Mean,
x = max(krill_base_SSS$year),
y = max(krill_base_SSS$SSS),
PF_text = glue("{round(PD, 3)}")
)
p_base_sss <- krill_base_SSS |>
ggplot(aes(x = year, y = SSS, group = sim)) +
geom_path(
data = ~filter(.x, below_ref == FALSE),
alpha = 0.3,
color = "#7CB342",
linewidth = 0.3
) +
geom_path(
data = ~filter(.x, below_ref == TRUE),
alpha = 0.5,
color = "#E53935",
linewidth = 0.5
) +
geom_hline(yintercept = kb_pms$PD@Ref, linetype = "dashed") +
geom_label(
data = krill_base_PD_mtc,
#aes(x = x, y = y, label = PF_text),
aes(x = 18, y = 3.75, label = PF_text),
#hjust = 1, vjust = 1,
size = 2.5, colour = "gray25",
inherit.aes = FALSE
) +
ggh4x::facet_wrap2(.~mp, ncol = 4, strip = MP_strips_theme) +
labs(
x = "Year",
y = expression(paste("Spawning stock status (SSB/", SSB[0], ")"))
)
ggsave(
plot = p_base_sss,
filename = "outputs/plots/fig_krill_mse_base_SSS_trajectories.png",
width = 8,
height = 10
)
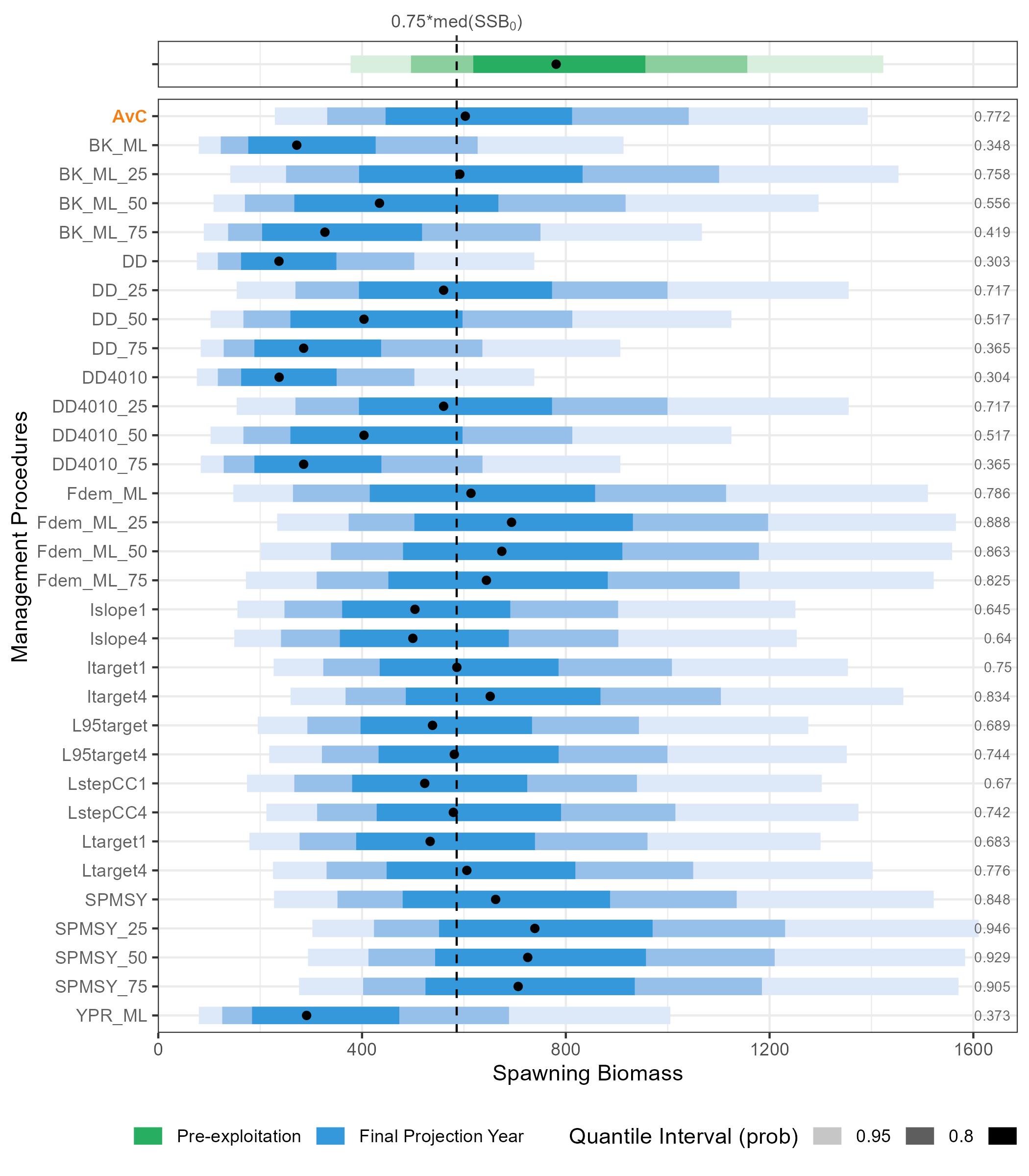
Virgin Spawning Biomass Vs Spawning Biomass in last Projection Year
Code
krill_base_SSBY <- read_rds("outputs/PM_results/krill_base_SSBY.rds") |>
reshape2::melt( varnames = c("sim", "mp_idx")) |>
dplyr::left_join(MP_idx2name, by = "mp_idx") |>
mutate(ssb_type = "Final Projection Year")
krill_base_SSB0 <- tibble(
value = read_rds("outputs/PM_results/krill_base_SSB0.rds") ,
sim = 1:length(value),
mp = "",
ssb_type = "Pre-exploitation")
krill_base_ESC_mtc <- tibble(
mp = kb_pms$ESC@MPs,
ESC = kb_pms$ESC@Mean,
ESC_text = glue("{round(ESC, 3)}"),
ssb_type = "Final Projection Year") |>
mutate(ssb_type = fct_relevel(ssb_type, "Pre-exploitation"))
x_text_col <- ifelse(kb_pms$ESC@MPs == "AvC", col_status_quo, "#616161")
x_text_face <- ifelse(kb_pms$ESC@MPs == "AvC", "bold", "plain")
p_base_ssb0_ssby <- krill_base_SSBY |>
add_row(krill_base_SSB0) |>
mutate(ssb_type = fct_relevel(ssb_type, "Pre-exploitation")) |>
ggplot(aes(y = fct_relevel(mp, rev), x = value, color = ssb_type)) +
ggdist::stat_interval(aes(color_ramp = after_stat(level))) +
stat_summary(geom = "point", fun = "median", col = "black") +
geom_vline(aes(xintercept = median(krill_base_SSB0$value)*0.75), linetype = "dashed") +
geom_text(data = krill_base_ESC_mtc, aes(x = Inf, y = mp, label = ESC_text), size = 2.5, hjust = 1.2, col = "gray40") +
labs(x = "Spawning Biomass", y = "Management Procedures") +
theme(legend.position="bottom") +
facet_wrap(~ssb_type, strip.position = "right", ncol = 1, scales = "free_y") +
force_panelsizes(rows = c(0.1, 2)) +
scale_colour_ramp_discrete(name = "Quantile Interval (prob)")+
scale_colour_discrete(name = "", type = c("#27AE60", "#3498DB")) +
guides(
# emphasize AvC
y = guide_axis_manual(
label_colour = rev(x_text_col),
label_face = rev(x_text_face)
),
x.sec = guide_axis_manual(
breaks = median(krill_base_SSB0$value)*0.75,
labels = expression(paste("0.75*med(", SSB[0], ")")), label_color = "gray30"
)
) +
theme(
#remove facet_wrap panels
strip.background = element_blank(),
strip.text = element_blank()
)
ggsave(
plot = p_base_ssb0_ssby,
filename = "outputs/plots/fig_krill_mse_base_SSS0_vs_SSBY.png",
width = 7,
height = 8
)
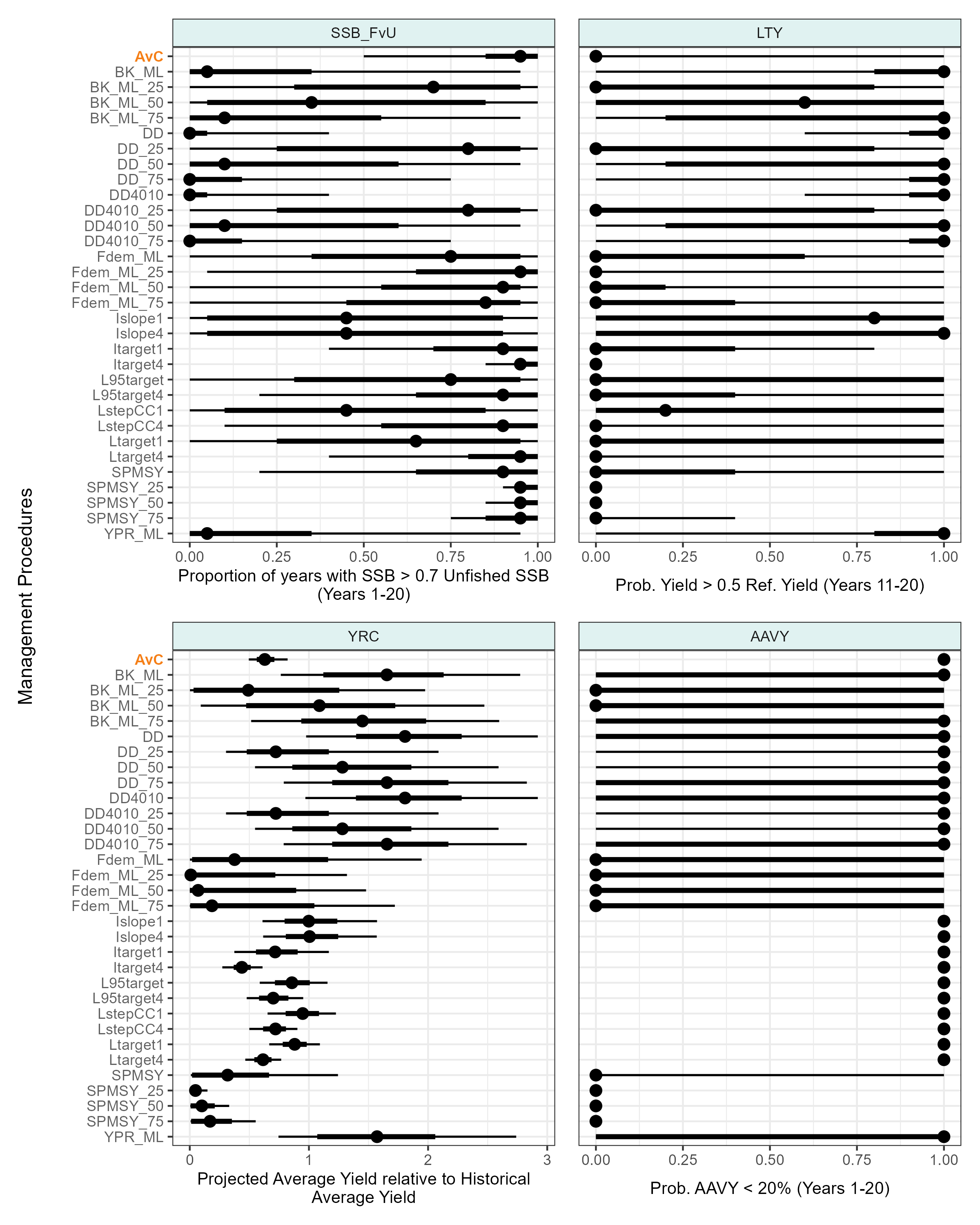
Fished Vs Unfished Spawning Biomass
Code
kb_pms_subset <- kb_pms[c("SSB_FvU", "LTY", "YRC", "AAVY")] |>
imap(\(x, y){
if(y == "YRC"){
x@Stat |>
reshape2::melt(
varnames = c("sim", "mp_idx")
) |>
dplyr::left_join(MP_idx2name, by = "mp_idx") |>
mutate(
pm = y,
caption = str_wrap(x@Name, 50)
)
}else{
x@Prob |>
reshape2::melt(
varnames = c("sim", "mp_idx")
) |>
dplyr::left_join(MP_idx2name, by = "mp_idx") |>
mutate(pm = y, caption = str_wrap(x@Caption, 50))
}
})
kb_pms_subset_plots <- kb_pms_subset |>
imap(\(x, y){
if(y %in% c("SSB_FvU", "YRC")){
y_text_col <- ifelse(unique(x$mp) == "AvC", col_status_quo, "#616161")
y_text_face <- ifelse(unique(x$mp) == "AvC", "bold", "plain")
p <- x |>
ggplot(aes(y = fct_relevel(mp, rev), x = value)) +
stat_pointinterval() +
# stat_interval() +
# stat_summary(geom = "point", fun = "median", col = "black") +
# scale_color_brewer() +
facet_wrap(~pm) +
labs(x = unique(x$caption), y = "") +
guides(
# emphasize AvC
y = guide_axis_manual(
label_colour = rev(y_text_col),
label_face = rev(y_text_face)
)
)
}else{
p <- x |>
ggplot(aes(y = fct_relevel(mp, rev), x = value)) +
stat_pointinterval() +
# stat_interval() +
# stat_summary(geom = "point", fun = "median", col = "black") +
# scale_color_brewer() +
facet_wrap(~pm) +
labs(x = unique(x$caption)) +
theme(
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank()
)
}
p + theme(axis.title.x = element_text(size = 10))
})
# Y-axis title-only plot
y_title <- ggplot(data.frame(l = "Management Procedures", x = 1, y = 1)) +
geom_text(aes(x, y, label = l), angle = 90, size = 4) +
theme_void() +
coord_cartesian(clip = "off")
p_base_pms_sub <- y_title + (wrap_plots(kb_pms_subset_plots, nrow = 2)) + plot_layout(widths = c(1, 25))
ggsave(
plot = p_base_pms_sub,
filename = "outputs/plots/fig_krill_mse_base_draws_PMs_subset.png",
width = 8,
height = 10
)
Trade-off plots
Code
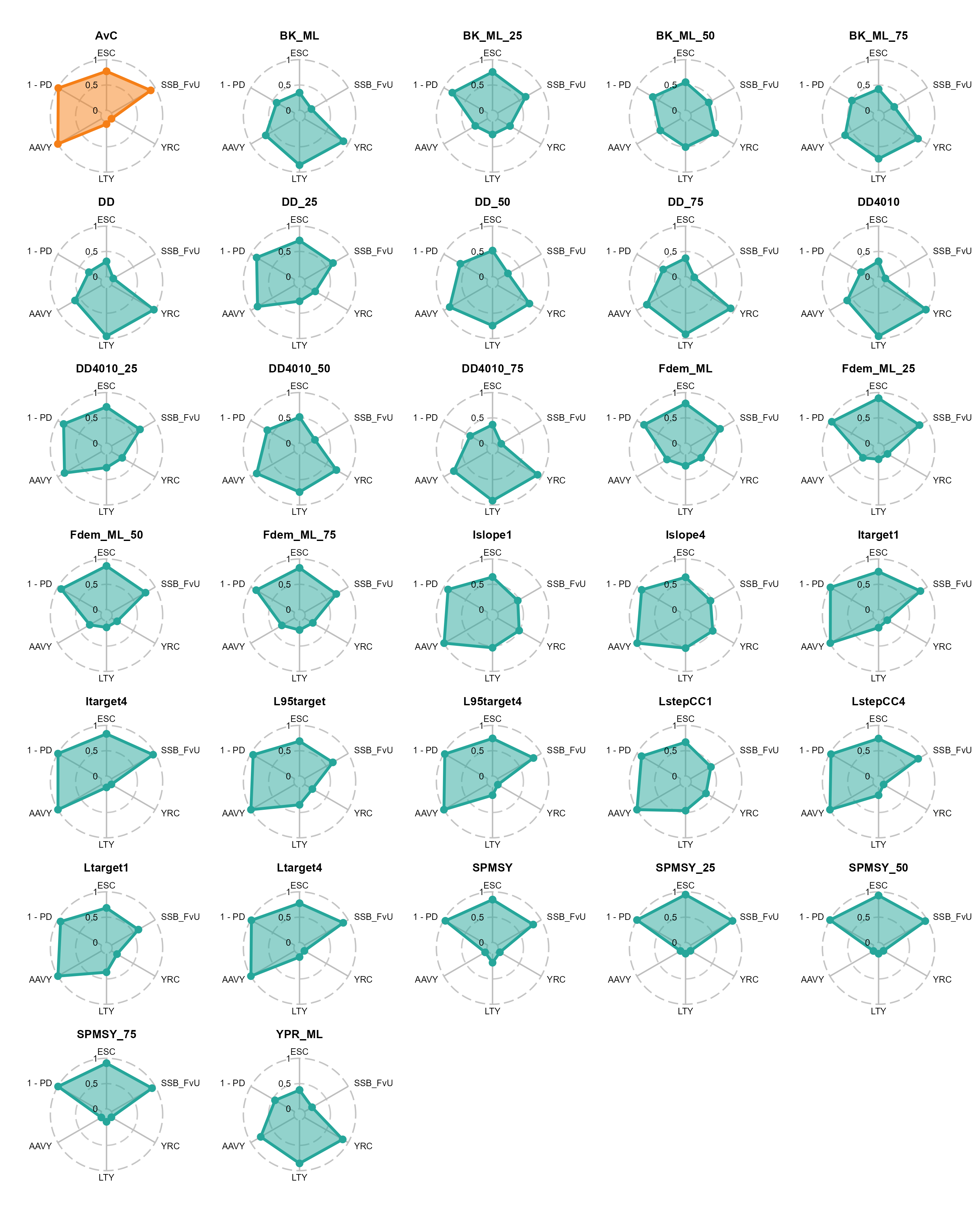
radars_base_pms <- kb_pms_summ |>
split(f = ~MP) |>
map(\(x){
x |>
dplyr::select(-PD) |>
relocate(`1 - PD`, .after = AAVY) |>
ggradar::ggradar(
values.radar = c(0, 0.5, 1),
axis.label.offset = 1.12,
# Polygons
group.line.width = 1,
group.point.size = 2,
group.colours = ifelse(x$MP == "AvC", col_status_quo, "#26A69A"),
# Background
background.circle.colour = "white",
#grid lines
gridline.label.offset = -.15 * (1 - (0 - ((1/9) * (1 - 0)))),
gridline.mid.colour = "grey",
# text size
axis.label.size = 2.3,
grid.label.size = 3,
# general stuff
fill = TRUE,
plot.title = x$MP
) +
theme(
plot.title = element_text(
size = 8.5, face = "bold", hjust = 0.5, #vjust = -1,
margin = margin(0, 0, 0, 0)
),
plot.margin = margin(1,1,1,1)
)
})
radars_base_pms_export <- wrap_plots(radars_base_pms, ncol = 5)
ggsave(
plot = radars_base_pms_export,
filename = "outputs/plots/fig_krill_mse_base_PMs_radars.png",
width = 10,
height = 12.5
)
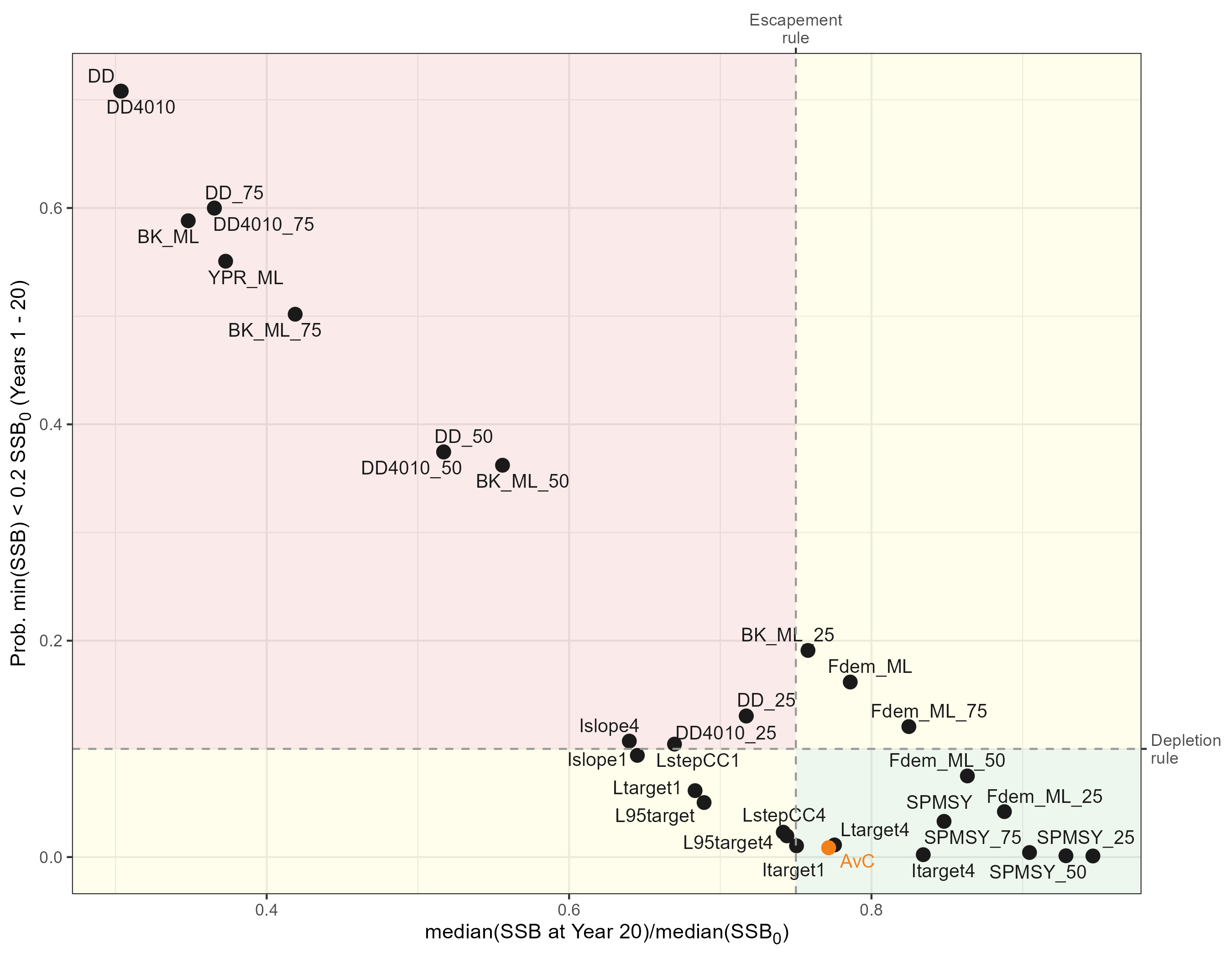
Satisficing CCAMLR’s Precautionary Rules
Code
ccamlr_pd_lim <- 0.1
ccamlr_esc_lim <- 0.75
x_label_pd <- str_replace_all(kb_pms$ESC@Caption, "SSB0", "SSB<sub>0</sub>")
y_label_esc <- str_replace_all(kb_pms$PD@Caption, "SSB0", "SSB<sub>0</sub>")
# Define satisficing areas in plot with associated color shading
# - fail both PD and ESC rules => top-left => red
# - satisfy PD but not ESC => bottom-left => yellow
# - satisfy ESC but not PP => top-right => yellow
# - satisfy both ESC and PP => bottom-right => green
cammlr_areas <- expand_grid(
position = c("top", "bottom"),
side = c("left", "right")
) |>
mutate(
area = glue("{position}-{side}"),
xmin = ifelse(str_detect(area, "left"), -Inf, ccamlr_esc_lim),
xmax = ifelse(str_detect(area, "left"), ccamlr_esc_lim, Inf),
ymin = ifelse(str_detect(area, "top"), ccamlr_pd_lim, -Inf),
ymax = ifelse(str_detect(area, "top"), Inf, ccamlr_pd_lim),
col = case_when(
area == "bottom-right" ~ "#4CAF50",
area == "top-left" ~ "#D32F2F",
area %in% c("top-right", "bottom-left") ~ "#FFEB3B"
)
)
p_satisficing <- kb_pms_summ |>
select(MP, PD, ESC) |>
mutate(is_status_quo = ifelse(MP == "AvC", "yes", "no")) |>
ggplot(aes(x = ESC, y = PD, label = MP, col = is_status_quo)) +
geom_rect(
data = cammlr_areas,
aes(xmin = xmin, xmax = xmax, ymin = ymin, ymax = ymax, fill = col),
alpha = 0.1,
inherit.aes = FALSE
) +
geom_point(size = 3) +
geom_point(data = ~filter(.x, MP == "AvC"), size = 3) +
geom_text_repel(size = 3.5) +
geom_vline(xintercept = ccamlr_esc_lim, linetype = "dashed", col = "gray60") +
geom_hline(yintercept = ccamlr_pd_lim, linetype = "dashed", col = "gray60") +
scale_fill_identity() +
scale_color_manual(values = c("gray10", col_status_quo)) +
labs(x = x_label_pd, y = y_label_esc) +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.position = "none"
) +
guides(
y.sec = guide_axis_manual(
breaks = 0.1,
labels = "Depletion\nrule"),
x.sec = guide_axis_manual(
breaks = 0.75,
labels = "Escapement\nrule")
)
ggsave(
plot = p_satisficing,
filename = "outputs/plots/fig_krill_mse_base_ccamlr_satisficing.png",
width = 9,
height = 7
)
4.4.1 Robustness Results
Code
# Run PM calculations on each robustness scenario
krill_rbst_pm <- dir_ls(path = "outputs/", regexp = "_rbst_") |>
map(\(mse_rbst_path){
mseobj <- read_rds(mse_rbst_path)
list(
PD = PD(mseobj, Ref = 0.2),
ESC = ESC(mseobj),
SSB_FvU = SSB_FvU(mseobj, Ref = 0.70),
YRC = YRC(mseobj),
LTY = LTY(mseobj),
AAVY = AAVY(mseobj)
)
},
.progress = TRUE)
names(krill_rbst_pm) <- str_extract(path_ext_remove(names(krill_rbst_pm)), "(?<=_rbst_)\\w+")
# store outputs
write_rds(krill_rbst_pm, "outputs/PM_results/krill_robustness_PMs_outputs.rds", "gz")Code
krill_rbst_pm <- read_rds("outputs/PM_results/krill_robustness_PMs_outputs.rds")
# Extract PM summary data
krill_rbst_pm_sums <- krill_rbst_pm |>
# iterate over robustness scenarios
imap(\(rbst_pms_out, rbst_name){
rbst_pms_out |>
# iterate over PM metrics
imap(\(pm_out, pm_name){
tibble(
MP = pm_out@MPs,
{{pm_name}} := pm_out@Mean
)
}) |>
reduce(full_join, by = "MP") |>
mutate(
#scn = paste0("rbst_", rbst_name),
scn = rbst_name,
scn_group = "Robustness\nScenarios",
.before = 1)
}) |>
list_rbind()
# Best MPs based on satisficing both CCAMLR rules (ESC and PD)
base_best_mps <- kb_pms_summ |>
select(-`1 - PD`) |>
filter(PD < 0.1 & ESC > 0.75) |>
mutate(scn = "base", scn_group = "Base\ncase")Code
# ------------------------------------------------------------------------------
# helper function for tabulating
# set icons setting trend of robustness scenarios relative to base-case, for each considered MP and PM
trend_indicator <- function(x, ref){
case_when(
x < ref ~ "images/caret-down.png",
x > ref ~ "images/caret-up.png",
.default = "images/minus.png"
)
}
satisfy_pd_rule <- function(x){
ifelse(x > 0.1, "#FFCDD2", "transparent")
}
satisfy_esc_rule <- function(x){
ifelse(x < 0.75, "#FFCDD2", "transparent")
}
# ---------------------------------------------------------------
# Prepare data for tabulating
krill_rbst_tbl <- krill_rbst_pm_sums |>
filter(MP %in% base_best_mps$MP) |>
bind_rows(base_best_mps) |>
select(-c(scn_group, SSB_FvU:AAVY)) |>
pivot_longer(cols = -c(scn, MP), names_to = "PM") |>
pivot_wider(names_from = scn, values_from = value) |>
relocate(base, .after = PM) |>
mutate(across(c(beta:qinc), ~trend_indicator(.x, base), .names = "{.col}_trend"))
key_cols <- str_subset(names(krill_rbst_tbl), "trend", negate = TRUE)
krill_rbst_tbl |>
flextable(
col_keys = key_cols
) |>
add_header_row(
values = c("Management Procedure", "Performance Metric", "Base Case", "Robustness Scenarios"),
colwidths = c(1, 1, 1, 5)) |>
align(align = "center", part = "all") |>
set_header_labels(
MP = "Management Procedure",
PM = "Performance Metric",
base = "Base Case",
beta = "Hyperstability",
qinc = "Increasing Fishing Efficiency",
LenCV = "Higher CV in Length-at-age",
Cbiascv = "Bias in Catch",
Mbiascv = "Bias in Natural Mortality"
) |>
merge_v( j = 1:3, part = "header", ) |>
merge_v(j = ~ MP) |>
colformat_double() |>
append_chunks(j = ~beta, as_chunk(" "), as_image(src = beta_trend, width = .15, height = .2)) |>
append_chunks(j = ~Cbiascv, as_chunk(" "), as_image(src = Cbiascv_trend, width = .15, height = .2)) |>
append_chunks(j = ~LenCV, as_chunk(" "), as_image(src = LenCV_trend, width = .15, height = .2)) |>
append_chunks(j = ~Mbiascv, as_chunk(" "), as_image(src = Mbiascv_trend, width = .15, height = .2)) |>
append_chunks(j = ~qinc, as_chunk(" "), as_image(src = qinc_trend, width = .15, height = .2)) |>
highlight(i = ~ PM %in% "PD", j = ~beta + Cbiascv + LenCV + Mbiascv + qinc , color = satisfy_pd_rule) |>
highlight(i = ~ PM %in% "ESC", j = ~beta + Cbiascv + LenCV + Mbiascv + qinc, color = satisfy_esc_rule) |>
bg(j = ~ base, bg = "#F5F5F5", part = "all") |>
fix_border_issues()Management Procedure | Performance Metric | Base Case | Robustness Scenarios | ||||
|---|---|---|---|---|---|---|---|
Hyperstability | Bias in Catch | Higher CV in Length-at-age | Bias in Natural Mortality | Increasing Fishing Efficiency | |||
AvC | PD | 0.0086 | 0.0086 | 0.0088 | 0.0076 | 0.0086 | 0.0086 |
ESC | 0.7717 | 0.7717 | 0.7725 | 0.7751 | 0.7717 | 0.7717 | |
Itarget1 | PD | 0.0104 | 0.0098 | 0.0102 | 0.0090 | 0.0104 | 0.0104 |
ESC | 0.7504 | 0.7513 | 0.7491 | 0.7538 | 0.7504 | 0.7504 | |
Itarget4 | PD | 0.0022 | 0.0022 | 0.0022 | 0.0020 | 0.0022 | 0.0022 |
ESC | 0.8343 | 0.8352 | 0.8354 | 0.8377 | 0.8343 | 0.8343 | |
Ltarget4 | PD | 0.0112 | 0.0112 | 0.0118 | 0.0102 | 0.0108 | 0.0112 |
ESC | 0.7756 | 0.7759 | 0.7779 | 0.7810 | 0.7759 | 0.7756 | |
Fdem_ML_25 | PD | 0.0420 | 0.0428 | 0.0432 | 0.0578 | 0.0422 | 0.0420 |
ESC | 0.8879 | 0.8891 | 0.8900 | 0.8739 | 0.8919 | 0.8879 | |
Fdem_ML_50 | PD | 0.0750 | 0.0716 | 0.0782 | 0.1170 | 0.0788 | 0.0750 |
ESC | 0.8634 | 0.8639 | 0.8626 | 0.8294 | 0.8651 | 0.8634 | |
SPMSY | PD | 0.0330 | 0.0412 | 0.0378 | 0.0354 | 0.0380 | 0.0330 |
ESC | 0.8480 | 0.8454 | 0.8397 | 0.8514 | 0.8363 | 0.8480 | |
SPMSY_25 | PD | 0.0010 | 0.0010 | 0.0010 | 0.0008 | 0.0012 | 0.0010 |
ESC | 0.9465 | 0.9466 | 0.9442 | 0.9439 | 0.9483 | 0.9465 | |
SPMSY_50 | PD | 0.0012 | 0.0016 | 0.0014 | 0.0010 | 0.0010 | 0.0012 |
ESC | 0.9286 | 0.9273 | 0.9268 | 0.9288 | 0.9249 | 0.9286 | |
SPMSY_75 | PD | 0.0042 | 0.0042 | 0.0040 | 0.0036 | 0.0040 | 0.0042 |
ESC | 0.9045 | 0.9025 | 0.9015 | 0.9073 | 0.9031 | 0.9045 | |
Graphical alternative to above table.
Code
krill_rbst_pm_sums_best <- krill_rbst_pm_sums |>
filter(MP %in% base_best_mps$MP)
krill_rbst_pm_sums_best <- left_join(krill_rbst_pm_sums_best, rbstn_tbl |> select(c(`Robustness Test ID`, `OM Parameter`)), by = c("scn" = "OM Parameter")) |>
select(-scn) |>
rename(scn = "Robustness Test ID")
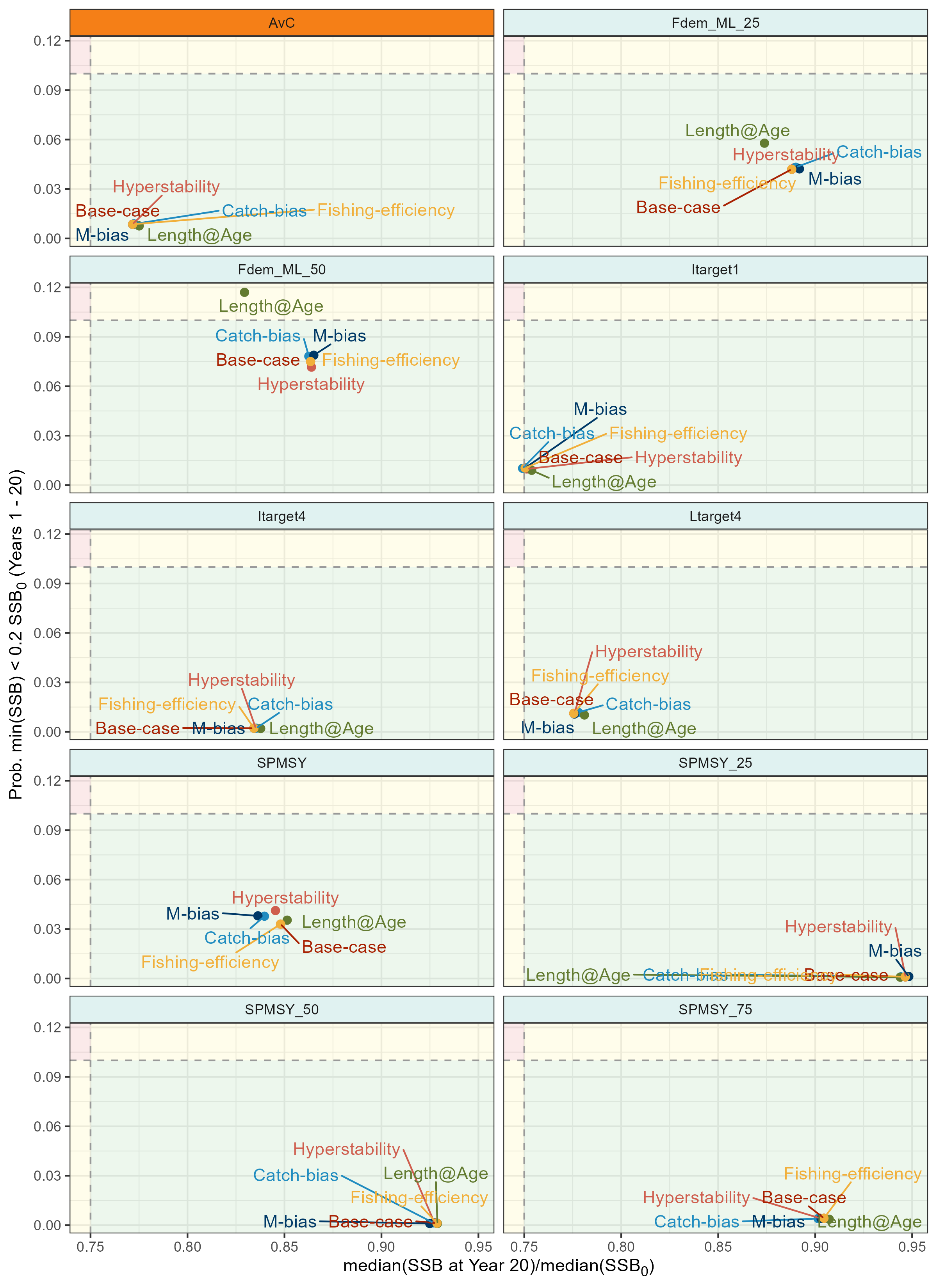
p_robustness <- bind_rows(base_best_mps, krill_rbst_pm_sums_best) |>
select(scn, MP, PD, ESC, scn_group) |>
mutate(scn = ifelse(scn == "base", "Base-case", scn)) |>
ggplot() +
geom_rect(
data = cammlr_areas,
aes(xmin = xmin, xmax = xmax, ymin = ymin, ymax = ymax, fill = col),
alpha = 0.1,
inherit.aes = FALSE
) +
geom_point(aes(x = ESC, y = PD, col = scn), size = 2) +
geom_text_repel(
aes(x = ESC, y = PD, col = scn, label = scn),
max.overlaps = 15,
#fontface = "bold",
family = "sans"
) +
geom_vline(xintercept = ccamlr_esc_lim, linetype = "dashed", col = "gray60") +
geom_hline(yintercept = ccamlr_pd_lim, linetype = "dashed", col = "gray60") +
facet_wrap2(vars(MP), ncol = 2, strip = MP_strips_theme) +
labs(x = x_label_pd, y = y_label_esc) +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.position = "none"
) +
scale_fill_identity() +
#scale_color_met_d("Moreau") ++
scale_color_met_d("Juarez") +
guides(colour="none")
#p_robustness
ggsave(
plot = p_robustness,
filename = "outputs/plots/fig_krill_mse_robustness.png",
width = 8,
height = 11
)
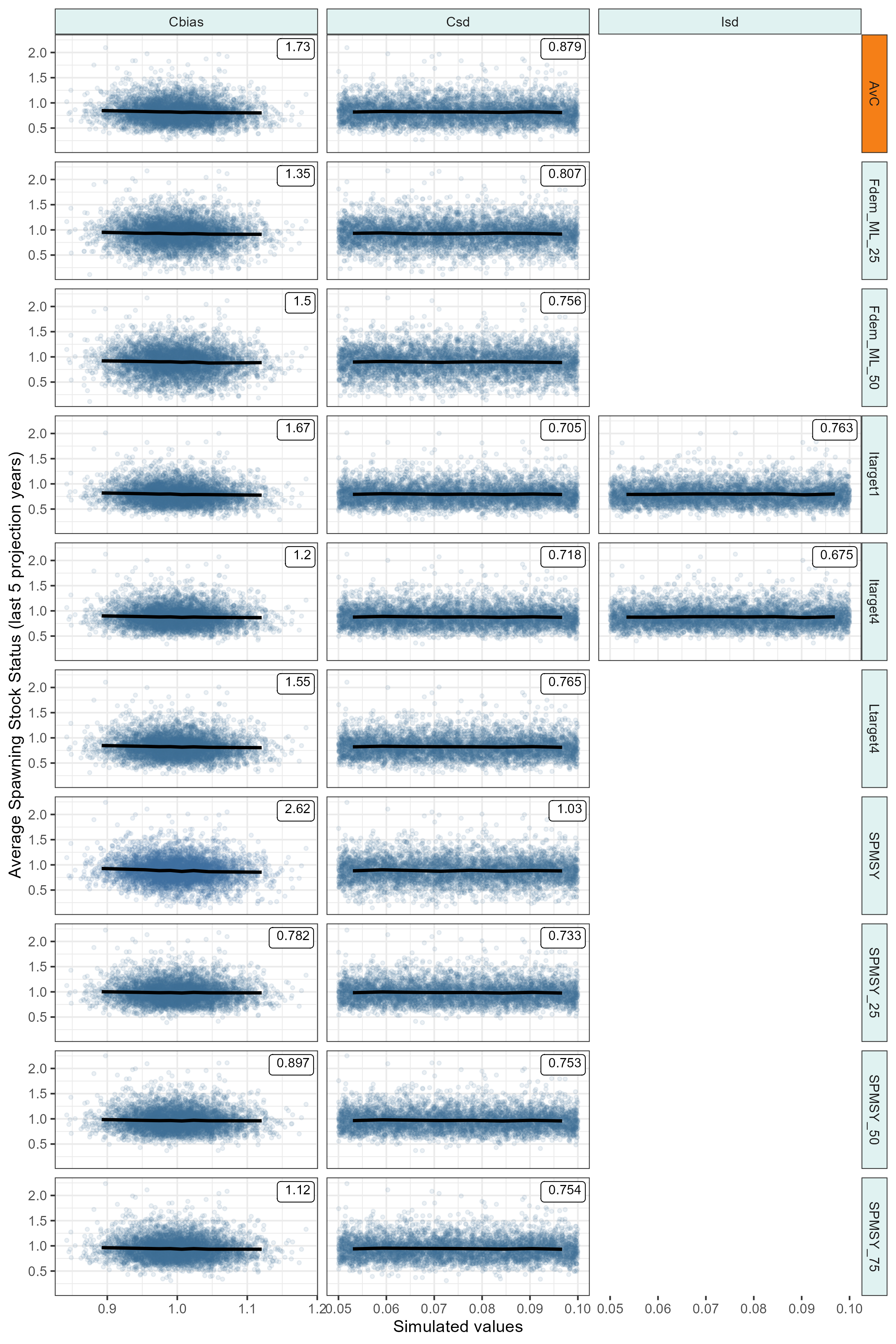
Value of Information plots
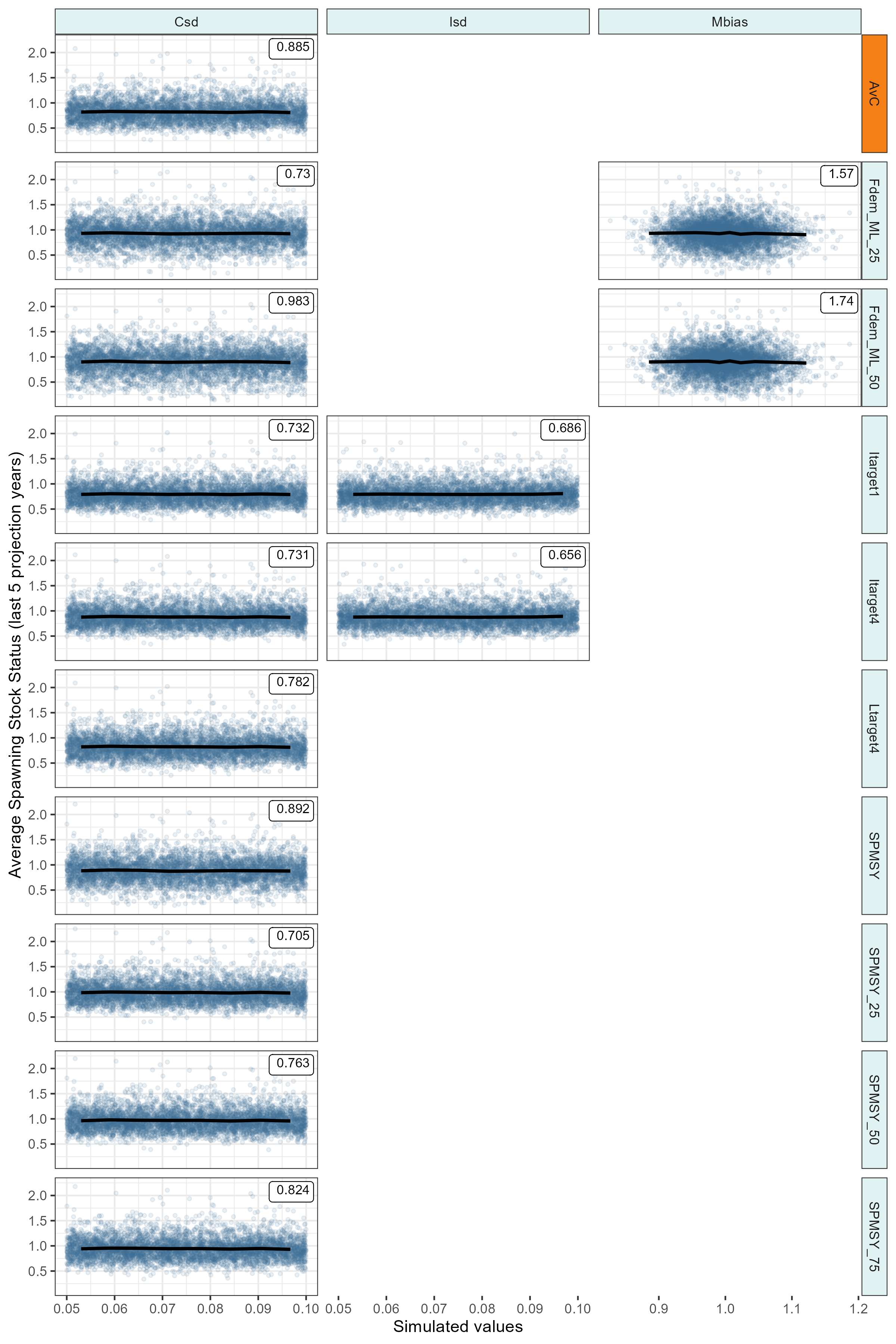
Exploring the sensitivity of Management Procedures to variability in the observation process.
Code
source("helpers/VOI_hack.r")
# run VOI calculations for robustness scenarios accounting for observation bias
krill_rbst_voi <- dir_ls(path = "outputs/", regexp = "bias") |>
map(\(mse_rbst_path){
mseobj <- read_rds(mse_rbst_path)
mseobj_best <- Sub(mseobj, MPs = base_best_mps$MP)
# calculate average SSS in last 5 years for each simulation (row) and MP (column)
SSS <- apply(PD(mseobj_best, Yrs = -5)@Stat, c(1, 2), mean)
VOI_hack(
mseobj_best,
Ut = SSS,
Utnam = "Average Spawning Stock Status (last 5 projection years)",
plot = TRUE,
maxrow = 10,
ref_MP = "AvC", col_refMP = col_status_quo, col_otherMP = col_facet_strip_bg
)
},
.progress = TRUE)
names(krill_rbst_voi) <- str_extract(path_ext_remove(names(krill_rbst_voi)), "(?<=_rbst_)\\w+")
krill_rbst_voi |>
iwalk(\(x, y){
ggsave(
plot = x$p,
filename = paste0("outputs/plots/fig_krill_robustness_voi_", y, ".png"),
width = 8,
height = 12
)
})
4.5 Session Info
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31 ucrt)
os Windows 11 x64 (build 22621)
system x86_64, mingw32
ui RTerm
language (EN)
collate English_United Kingdom.utf8
ctype English_United Kingdom.utf8
tz Europe/London
date 2023-12-20
pandoc 3.1.1 @ C:/Program Files/RStudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P abind 1.4-5 2016-07-21 [?] CRAN (R 4.3.0)
P askpass 1.1 2019-01-13 [?] CRAN (R 4.3.1)
P bit 4.0.5 2022-11-15 [?] CRAN (R 4.3.1)
P bit64 4.0.5 2020-08-30 [?] CRAN (R 4.3.1)
P bitops 1.0-7 2021-04-24 [?] CRAN (R 4.3.0)
P cachem 1.0.8 2023-05-01 [?] CRAN (R 4.3.1)
P caTools 1.18.2 2021-03-28 [?] CRAN (R 4.3.1)
P cellranger 1.1.0 2016-07-27 [?] CRAN (R 4.3.1)
P cli * 3.6.1 2023-03-23 [?] CRAN (R 4.3.1)
P codetools 0.2-19 2023-02-01 [?] CRAN (R 4.3.2)
P colorspace 2.1-0 2023-01-23 [?] CRAN (R 4.3.1)
P commonmark 1.9.0 2023-03-17 [?] CRAN (R 4.3.1)
P crayon 1.5.2 2022-09-29 [?] CRAN (R 4.3.1)
P crul 1.4.0 2023-05-17 [?] CRAN (R 4.3.1)
P curl 5.0.2 2023-08-14 [?] CRAN (R 4.3.1)
P data.table 1.14.8 2023-02-17 [?] CRAN (R 4.3.1)
P devtools 2.4.5 2022-10-11 [?] CRAN (R 4.3.2)
P digest 0.6.33 2023-07-07 [?] CRAN (R 4.3.1)
P distributional 0.3.2 2023-03-22 [?] CRAN (R 4.3.1)
P DLMtool * 6.0.6 2022-06-20 [?] CRAN (R 4.3.1)
P dplyr * 1.1.2 2023-04-20 [?] CRAN (R 4.3.1)
P dtplyr * 1.3.1 2023-03-22 [?] CRAN (R 4.3.1)
P ellipsis 0.3.2 2021-04-29 [?] CRAN (R 4.3.1)
P equatags 0.2.0 2022-06-13 [?] CRAN (R 4.3.1)
P evaluate 0.21 2023-05-05 [?] CRAN (R 4.3.1)
P fansi 1.0.4 2023-01-22 [?] CRAN (R 4.3.1)
P farver 2.1.1 2022-07-06 [?] CRAN (R 4.3.1)
P fastmap 1.1.1 2023-02-24 [?] CRAN (R 4.3.1)
P flextable * 0.9.2 2023-06-18 [?] CRAN (R 4.3.1)
P fontBitstreamVera 0.1.1 2017-02-01 [?] CRAN (R 4.3.0)
P fontLiberation 0.1.0 2016-10-15 [?] CRAN (R 4.3.0)
P fontquiver 0.2.1 2017-02-01 [?] CRAN (R 4.3.1)
P forcats * 1.0.0 2023-01-29 [?] CRAN (R 4.3.1)
P fs * 1.6.3 2023-07-20 [?] CRAN (R 4.3.1)
P furrr * 0.3.1 2022-08-15 [?] CRAN (R 4.3.1)
P future * 1.33.0 2023-07-01 [?] CRAN (R 4.3.1)
P gdtools 0.3.3 2023-03-27 [?] CRAN (R 4.3.1)
P generics 0.1.3 2022-07-05 [?] CRAN (R 4.3.1)
P gfonts 0.2.0 2023-01-08 [?] CRAN (R 4.3.1)
P ggblend * 0.1.0 2023-05-22 [?] CRAN (R 4.3.1)
P ggdensity * 1.0.0 2023-02-09 [?] CRAN (R 4.3.1)
P ggdist * 3.3.0 2023-05-13 [?] CRAN (R 4.3.1)
P ggh4x * 0.2.6 2023-08-30 [?] CRAN (R 4.3.2)
P ggplot2 * 3.4.3 2023-08-14 [?] CRAN (R 4.3.1)
ggradar * 0.2 2023-12-12 [1] Github (ricardo-bion/ggradar@53404a5)
P ggrepel * 0.9.4 2023-10-13 [?] CRAN (R 4.3.2)
P ggtext * 0.1.2 2022-09-16 [?] CRAN (R 4.3.2)
P globals 0.16.2 2022-11-21 [?] CRAN (R 4.3.0)
P glue * 1.6.2 2022-02-24 [?] CRAN (R 4.3.1)
P gplots 3.1.3 2022-04-25 [?] CRAN (R 4.3.1)
P gridtext 0.1.5 2022-09-16 [?] CRAN (R 4.3.2)
Grym * 0.1.1 2023-08-28 [1] Github (AustralianAntarcticDivision/Grym@6a2d6a1)
P gtable 0.3.4 2023-08-21 [?] CRAN (R 4.3.1)
P gtools 3.9.4 2022-11-27 [?] CRAN (R 4.3.1)
P hms 1.1.3 2023-03-21 [?] CRAN (R 4.3.1)
P htmltools 0.5.7 2023-11-03 [?] CRAN (R 4.3.2)
P htmlwidgets 1.6.4 2023-12-06 [?] CRAN (R 4.3.2)
P httpcode 0.3.0 2020-04-10 [?] CRAN (R 4.3.1)
P httpuv 1.6.11 2023-05-11 [?] CRAN (R 4.3.1)
P jsonlite 1.8.7 2023-06-29 [?] CRAN (R 4.3.1)
P katex 1.4.1 2022-11-28 [?] CRAN (R 4.3.1)
P KernSmooth 2.23-22 2023-07-10 [?] CRAN (R 4.3.2)
P knitr 1.43 2023-05-25 [?] CRAN (R 4.3.1)
P labeling 0.4.2 2020-10-20 [?] CRAN (R 4.3.0)
P later 1.3.1 2023-05-02 [?] CRAN (R 4.3.1)
P lattice 0.21-9 2023-10-01 [?] CRAN (R 4.3.2)
P lifecycle 1.0.3 2022-10-07 [?] CRAN (R 4.3.1)
P listenv 0.9.0 2022-12-16 [?] CRAN (R 4.3.1)
P lmtest 0.9-40 2022-03-21 [?] CRAN (R 4.3.1)
P lubridate * 1.9.2 2023-02-10 [?] CRAN (R 4.3.1)
P magrittr 2.0.3 2022-03-30 [?] CRAN (R 4.3.1)
P markdown 1.12 2023-12-06 [?] CRAN (R 4.3.2)
P MASS 7.3-60 2023-05-04 [?] CRAN (R 4.3.2)
P Matrix 1.6-1.1 2023-09-18 [?] CRAN (R 4.3.2)
P memoise 2.0.1 2021-11-26 [?] CRAN (R 4.3.1)
P MetBrewer * 0.2.0 2022-03-21 [?] CRAN (R 4.3.1)
P mime 0.12 2021-09-28 [?] CRAN (R 4.3.0)
P miniUI 0.1.1.1 2018-05-18 [?] CRAN (R 4.3.2)
P MSEtool * 3.7.0 2023-07-19 [?] CRAN (R 4.3.1)
P munsell 0.5.0 2018-06-12 [?] CRAN (R 4.3.1)
P mvtnorm * 1.2-3 2023-08-25 [?] CRAN (R 4.3.1)
P nlme 3.1-163 2023-08-09 [?] CRAN (R 4.3.2)
P officer 0.6.2 2023-03-28 [?] CRAN (R 4.3.1)
P openMSE * 1.0.0 2021-02-08 [?] CRAN (R 4.3.1)
P openssl 2.1.0 2023-07-15 [?] CRAN (R 4.3.1)
P openxlsx * 4.2.5.2 2023-02-06 [?] CRAN (R 4.3.1)
P parallelly 1.36.0 2023-05-26 [?] CRAN (R 4.3.0)
P patchwork * 1.1.3 2023-08-14 [?] CRAN (R 4.3.1)
P pbapply 1.7-2 2023-06-27 [?] CRAN (R 4.3.1)
P pillar 1.9.0 2023-03-22 [?] CRAN (R 4.3.1)
P pkgbuild 1.4.3 2023-12-10 [?] CRAN (R 4.3.2)
P pkgconfig 2.0.3 2019-09-22 [?] CRAN (R 4.3.1)
P pkgload 1.3.3 2023-09-22 [?] CRAN (R 4.3.2)
P plyr 1.8.8 2022-11-11 [?] CRAN (R 4.3.1)
P profvis 0.3.8 2023-05-02 [?] CRAN (R 4.3.2)
P progressr * 0.14.0 2023-08-10 [?] CRAN (R 4.3.1)
P promises 1.2.1 2023-08-10 [?] CRAN (R 4.3.1)
P purrr * 1.0.2 2023-08-10 [?] CRAN (R 4.3.1)
P R6 2.5.1 2021-08-19 [?] CRAN (R 4.3.1)
P ragg 1.2.5 2023-01-12 [?] CRAN (R 4.3.1)
P Rcpp 1.0.11 2023-07-06 [?] CRAN (R 4.3.1)
PD RcppParallel 5.1.7 2023-02-27 [?] CRAN (R 4.3.2)
P RcppZiggurat 0.1.6 2020-10-20 [?] CRAN (R 4.3.2)
P readr * 2.1.4 2023-02-10 [?] CRAN (R 4.3.1)
P readxl * 1.4.3 2023-07-06 [?] CRAN (R 4.3.1)
P remotes 2.4.2.1 2023-07-18 [?] CRAN (R 4.3.1)
renv 0.17.0 2023-03-02 [1] CRAN (R 4.3.1)
P reshape2 1.4.4 2020-04-09 [?] CRAN (R 4.3.1)
P Rfast 2.1.0 2023-11-09 [?] CRAN (R 4.3.2)
P rlang * 1.1.1 2023-04-28 [?] CRAN (R 4.3.1)
P rmarkdown 2.24 2023-08-14 [?] CRAN (R 4.3.1)
P rstudioapi 0.15.0 2023-07-07 [?] CRAN (R 4.3.1)
P SAMtool * 1.6.1 2023-08-23 [?] CRAN (R 4.3.1)
P sandwich 3.0-2 2022-06-15 [?] CRAN (R 4.3.1)
P scales 1.2.1 2022-08-20 [?] CRAN (R 4.3.1)
P sessioninfo 1.2.2 2021-12-06 [?] CRAN (R 4.3.2)
P shiny 1.7.5 2023-08-12 [?] CRAN (R 4.3.1)
P snow * 0.4-4 2021-10-27 [?] CRAN (R 4.3.0)
P snowfall * 1.84-6.2 2022-07-05 [?] CRAN (R 4.3.0)
P stringi 1.7.12 2023-01-11 [?] CRAN (R 4.3.0)
P stringr * 1.5.0 2022-12-02 [?] CRAN (R 4.3.1)
P strucchange 1.5-3 2022-06-15 [?] CRAN (R 4.3.1)
P svglite * 2.1.2 2023-10-11 [?] CRAN (R 4.3.2)
P systemfonts 1.0.4 2022-02-11 [?] CRAN (R 4.3.1)
P textshaping 0.3.6 2021-10-13 [?] CRAN (R 4.3.1)
P tibble * 3.2.1 2023-03-20 [?] CRAN (R 4.3.1)
P tictoc * 1.2 2023-04-23 [?] CRAN (R 4.3.1)
P tidyr * 1.3.0 2023-01-24 [?] CRAN (R 4.3.1)
P tidyselect 1.2.0 2022-10-10 [?] CRAN (R 4.3.1)
P tidyverse * 2.0.0 2023-02-22 [?] CRAN (R 4.3.1)
P timechange 0.2.0 2023-01-11 [?] CRAN (R 4.3.1)
D TMB 1.9.6 2023-08-11 [1] CRAN (R 4.3.1)
P tzdb 0.4.0 2023-05-12 [?] CRAN (R 4.3.1)
P urca 1.3-3 2022-08-29 [?] CRAN (R 4.3.1)
P urlchecker 1.0.1 2021-11-30 [?] CRAN (R 4.3.2)
P usethis 2.2.2 2023-07-06 [?] CRAN (R 4.3.2)
P utf8 1.2.3 2023-01-31 [?] CRAN (R 4.3.1)
P uuid 1.1-1 2023-08-17 [?] CRAN (R 4.3.1)
P V8 4.3.3 2023-07-18 [?] CRAN (R 4.3.1)
P vars 1.5-9 2023-03-22 [?] CRAN (R 4.3.1)
P vctrs 0.6.3 2023-06-14 [?] CRAN (R 4.3.1)
P vroom 1.6.3 2023-04-28 [?] CRAN (R 4.3.1)
P withr 2.5.0 2022-03-03 [?] CRAN (R 4.3.1)
P xfun 0.40 2023-08-09 [?] CRAN (R 4.3.1)
P xml2 1.3.5 2023-07-06 [?] CRAN (R 4.3.1)
P xslt 1.4.4 2023-02-21 [?] CRAN (R 4.3.1)
P xtable 1.8-4 2019-04-21 [?] CRAN (R 4.3.1)
P yaml 2.3.7 2023-01-23 [?] CRAN (R 4.3.0)
P zip 2.3.0 2023-04-17 [?] CRAN (R 4.3.1)
P zoo 1.8-12 2023-04-13 [?] CRAN (R 4.3.1)
[1] C:/Users/bruno/Dropbox/krill/krill_mse/renv/library/R-4.3/x86_64-w64-mingw32
[2] C:/Users/bruno/AppData/Local/R/cache/R/renv/sandbox/R-4.3/x86_64-w64-mingw32/7df9739c
P ── Loaded and on-disk path mismatch.
D ── DLL MD5 mismatch, broken installation.
──────────────────────────────────────────────────────────────────────────────#library(gratitude)